Command Line Tools¶
The MAX-ARD SDK CLI tools have built-in capabilities for working with ARD data stored in an Amazon S3 location. For example, the max-ard storage describe command describes the data in the specified S3 bucket. This command can also be used to generate a QGIS layer definition of the files stored in S3.
See example requests that use max-ard storage describe.
In addition to the Maxar ARD SDK CLI tools, the following tools are useful for working with ARD imagery in cloud storage:
AWS Command Line Interface¶
Amazon's AWS CLI is a set of command line tools for interacting with AWS services. The AWS CLI can be used to access and work with ARD data stored in an S3 bucket.
Installation¶
To install AWS CLI, see the AWS CLI Installation Guide. Choose your operation system from the list.
Once you've installed the AWS CLI, you'll need to configure it to use your AWS credentials to access data in your S3 bucket. See the AWS CLI Configuration Guide for details.
Accessing the Maxar ARD public datasets¶
The datasets used in the examples below are publicly available and do not require authentication. However, AWS CLI still checks for a current token for access. If you get an ExpiredToken error while running any of the AWS CLI examples below, you can append --no-sign-request to the command to skip the check.
Example:
aws s3 ls maxar-ard-samples/sample-002/ --no-sign-request
A sample dataset over San Francisco can be downloaded from the Maxar website. You will be asked to fill out a contact form before accessing the data. Download Sample Data
Use AWS CLI commands to work with ARD data¶
aws s3 ls (list files)¶
The ls command lists the files and prefixes in the requested path. In this example, we want to list the contents of the maxar-ard-samples/sample-002/ location.
aws s3 ls maxar-ard-samples/sample-002/
PRE 13/
PRE acquisition_collections/
PRE order_collections/
2021-04-12 11:11:02 243 5633324896324598239-request.json
sample-002
Note: Listing the contents of a prefix requires the trailing slash / at the end of the prefix name. If the trailing slash is not included, the results will only show the name of the prefix in the request. In this case, it will show sample-002.
To list all files within the sample-002 prefix and its descendants, include --recursive in your request.
aws s3 ls maxar-ard-samples/sample-002/ --recursive
2021-04-12 19:02:12 98304 sample-002/13/031133320010/2020-07-29/10300100AA1C6800-cloud-mask.gpkg
2021-04-12 19:02:12 98304 sample-002/13/031133320010/2020-07-29/10300100AA1C6800-data-mask.gpkg
2021-04-12 19:02:20 71193652 sample-002/13/031133320010/2020-07-29/10300100AA1C6800-ms.tif
2021-04-12 19:02:15 135381264 sample-002/13/031133320010/2020-07-29/10300100AA1C6800-pan.tif
2021-04-12 19:02:12 61112300 sample-002/13/031133320010/2020-07-29/10300100AA1C6800-visual.tif
2021-04-12 19:02:23 2104 sample-002/13/031133320010/2020-07-29/10300100AA1C6800.json
2021-04-12 18:37:40 98304 sample-002/13/031133320010/2020-07-29/10300100AC94D700-cloud-mask.gpkg
2021-04-12 18:37:40 98304 sample-002/13/031133320010/2020-07-29/10300100AC94D700-data-mask.gpkg
2021-04-12 18:37:49 74875710 sample-002/13/031133320010/2020-07-29/10300100AC94D700-ms.tif
2021-04-12 18:37:43 142014484 sample-002/13/031133320010/2020-07-29/10300100AC94D700-pan.tif
2021-04-12 18:37:40 61868775 sample-002/13/031133320010/2020-07-29/10300100AC94D700-visual.tif
2021-04-12 18:37:51 2106 sample-002/13/031133320010/2020-07-29/10300100AC94D700.json
2021-04-12 18:36:36 1648 sample-002/acquisition_collections/10300100AC700900_collection.json
2021-04-12 18:38:09 4122 sample-002/acquisition_collections/10300100AC94D700_collection.json
2021-04-12 20:21:33 1078 sample-002/acquisition_collections/10300100ACCDAB00_collection.json
2021-04-12 18:37:44 1650 sample-002/acquisition_collections/10300100AD437400_collection.json
2021-04-12 15:13:26 2982 sample-002/acquisition_collections/10300100B2B49700_collection.json
2021-04-12 15:13:02 1648 sample-002/acquisition_collections/10300100B3841C00_collection.json
2021-04-12 15:13:27 2597 sample-002/acquisition_collections/10300100B39ACD00_collection.json
2021-02-26 14:40:48 2767 sample-002/order_collections/5600806342510769702_root_collection.json
2021-04-12 20:37:37 2749 sample-002/order_collections/5633324896324598239_root_collection.json
[results truncated]
Partial match filtering¶
Partial match filtering is performed automatically when an AWS CLI request is made. This is helpful when you only know part of the string, or when you want to find all files that match the partial path.
This example:
aws s3 ls maxar-ard-samples/sample-002/acquisition_collections/10300100
acquisition_collections prefix that start with 10300100.
2021-04-12 20:19:36 699 10300100A67D3100_collection.json
2021-04-12 20:18:47 1078 10300100A9547600_collection.json
2021-04-12 20:37:33 1268 10300100A9CC9200_collection.json
2021-04-12 19:02:32 3738 10300100AA1C6800_collection.json
2021-04-12 19:11:34 1647 10300100AB101A00_collection.json
2021-04-12 18:36:36 1648 10300100AC700900_collection.json
2021-04-12 18:38:09 4122 10300100AC94D700_collection.json
2021-04-12 20:21:33 1078 10300100ACCDAB00_collection.json
2021-04-12 18:37:44 1650 10300100AD437400_collection.json
2021-04-12 15:13:26 2982 10300100B2B49700_collection.json
2021-04-12 15:13:02 1648 10300100B3841C00_collection.json
2021-04-12 15:13:27 2597 10300100B39ACD00_collection.json
Find files that include a specific string¶
Use grep (linux, macOS) or findstr (windows) to find all of the files that include a specific string in the path.
Example: Find all files named visual.tif in the rio_deforestation dataset.
aws s3 ls maxar-ard-samples/rio_deforestation/ --recursive | grep "visual.tif"
2021-04-12 20:10:25 56654384 rio_deforestation/19/300022033202/2009-09-05/105041000218BB00-visual.tif
2021-04-12 20:10:15 28992496 rio_deforestation/19/300022033202/2010-07-01/1050410001B2E100-visual.tif
2021-04-12 18:48:08 56860968 rio_deforestation/19/300022033202/2011-05-11/103001000AC5B000-visual.tif
2021-04-12 19:21:27 66317737 rio_deforestation/19/300022033202/2011-08-12/103001000CB3FD00-visual.tif
2021-04-12 19:06:27 30197512 rio_deforestation/19/300022033202/2012-05-10/103001001826FC00-visual.tif
2021-04-12 17:57:07 29645545 rio_deforestation/19/300022033202/2012-09-04/1050410000818900-visual.tif
2021-04-12 19:10:00 58833258 rio_deforestation/19/300022033202/2013-05-09/1050410002473100-visual.tif
2021-04-12 19:09:01 55904842 rio_deforestation/19/300022033202/2013-08-17/10300100253F7900-visual.tif
2021-04-12 19:06:25 31344763 rio_deforestation/19/300022033202/2013-08-28/1030010026C30800-visual.tif
2021-04-12 19:11:53 34979004 rio_deforestation/19/300022033202/2013-12-23/10300100296ED400-visual.tif
2021-04-12 17:58:54 15285643 rio_deforestation/19/300022033202/2014-04-21/10504100105F9E00-visual.tif
2021-04-12 19:05:07 61825502 rio_deforestation/19/300022033202/2015-06-29/1030010044098C00-visual.tif
[truncated]
Example: List all files with acquisition ID "1050410000818900" in the path.
aws s3 ls maxar-ard-samples/rio_deforestation/ --recursive | grep "1050410000818900"
2021-04-12 17:57:07 98304 rio_deforestation/19/300022033202/2012-09-04/1050410000818900-cloud-mask.gpkg
2021-04-12 17:57:07 98304 rio_deforestation/19/300022033202/2012-09-04/1050410000818900-data-mask.gpkg
2021-04-12 17:57:14 35145697 rio_deforestation/19/300022033202/2012-09-04/1050410000818900-ms.tif
2021-04-12 17:57:09 128922686 rio_deforestation/19/300022033202/2012-09-04/1050410000818900-pan.tif
2021-04-12 17:57:07 29645545 rio_deforestation/19/300022033202/2012-09-04/1050410000818900-visual.tif
2021-04-12 17:57:15 2390 rio_deforestation/19/300022033202/2012-09-04/1050410000818900.json
2021-04-12 17:57:19 698 rio_deforestation/acquisition_collections/1050410000818900_collection.json
aws s3 cp (copy files)¶
The cp command copies files from s3 to a specified location. These examples show copying from the ARD sample data in S3 to a local directory.
Copy a specific file¶
This example copies the 1050410000818900.json file from its s3 location to the user's current local directory.
aws s3 cp s3://maxar-ard-samples/rio_deforestation/19/300022033202/2012-09-04/1050410000818900.json ./1050410000818900.json
download: s3://maxar-ard-samples/rio_deforestation/19/300022033202/2012-09-04/1050410000818900.json to ./1050410000818900.json
Copy all files in a prefix¶
This example shows copying all the files from the specified prefix to the user's current local directory using the --recursive option.
aws s3 cp s3://maxar-ard-samples/rio_deforestation/19/300022033202/2012-09-04/ ./ --recursive
download: s3://maxar-ard-samples/rio_deforestation/19/300022033202/2012-09-04/1050410000818900.json to ./1050410000818900.json
download: s3://maxar-ard-samples/rio_deforestation/19/300022033202/2012-09-04/1050410000818900-cloud-mask.gpkg to ./1050410000818900-cloud-mask.gpkg
download: s3://maxar-ard-samples/rio_deforestation/19/300022033202/2012-09-04/1050410000818900-data-mask.gpkg to ./1050410000818900-data-mask.gpkg
download: s3://maxar-ard-samples/rio_deforestation/19/300022033202/2012-09-04/1050410000818900-ms.tif to ./1050410000818900-ms.tif
download: s3://maxar-ard-samples/rio_deforestation/19/300022033202/2012-09-04/1050410000818900-visual.tif to ./1050410000818900-visual.tif
download: s3://maxar-ard-samples/rio_deforestation/19/300022033202/2012-09-04/1050410000818900-pan.tif to ./1050410000818900-pan.tif
aws s3 sync (synchronize files in two locations)¶
Use the sync command to synchronize the contents of two locations. This is typically used to keep files up to date. In this example, rio-deforestation is the source and rio-data is the target local directory.
aws s3 sync s3://maxar-ard-samples/rio_deforestation rio-data
Completed 2.1 KiB/~7.1 GiB (5.7 KiB/s) with ~146 file(s) remaining (calculating.download: s3://maxar-ard-samples/rio_deforestation/19/300022033202/2009-09-05/105041000218BB00.json to rio-data/19/300022033202/2009-09-05/105041000218BB00.json
Completed 2.1 KiB/~7.1 GiB (5.7 KiB/s) with ~145 file(s) remaining (calculating.Completed 98.1 KiB/~7.1 GiB (194.9 KiB/s) with ~146 file(s) remaining (calculatidownload: s3://maxar-ard-samples/rio_deforestation/19/300022033202/2009-09-05/105041000218BB00-cloud-mask.gpkg to rio-data/19/300022033202/2009-09-05/105041000218BB00-cloud-mask.gpkg
[truncated]
GDAL/OGR command line tools¶
Introduction¶
Files stored in S3 can be accessed using GDAL/OGR command line tools. Use gdalinfo to access raster data and ogrinfo to access vector data. For example, use gdalinfo to list information about a GeoTIFF, and 'ogrinfo' to list information about a cloud mask GeoPackage. Examples of both of these are shown below.
To access files stored in S3 using GDAl or OGR, use vsis3.
Installation¶
The GDAL/OGR CLI installs with GDAL. To find the latest release and download instructions, see GDAL Current Releases.
Use GDAL command line tools to work with ARD data in the cloud or in a local directory.¶
GDAL's vsis3 virtual driver can read files from S3. To use it, change the s3:// protocol in the file's URL to `/vsis3/:
| Protocol | File URL |
|---|---|
| S3 | s3://maxar-ard-samples/rio_deforestation/19/300022033202/2013-08-28/1030010026C30800-ms.tif |
| GDAL | /vsis3/maxar-ard-samples/rio_deforestation/19/300022033202/2013-08-28/1030010026C30800-ms.tif |
| local file | /[local dir/subdir]/19/300022033202/2013-08-28/1030010026C30800-ms.tif |
gdalinfo¶
This example lists information about the specified GeoTIFF (raster file).
gdalinfo /vsis3/maxar-ard-samples/rio_deforestation/19/300022033202/2013-08-28/1030010026C30800-ms.tif
Example using downloaded files from the sample dataset (SF)
gdalinfo /[root dir]/[subdir]/sample-001-subset/10/120020223032/2019-07-12/1050010017603F00-ms.tif
Driver: GTiff/GeoTIFF
Files: /vsis3/maxar-ard-samples/rio_deforestation/19/300022033202/2013-08-28/1030010026C30800-ms.tif
Size is 2383, 2383
Coordinate System is:
PROJCRS["WGS 84 / UTM zone 19S",
BASEGEOGCRS["WGS 84",
DATUM["World Geodetic System 1984",
ELLIPSOID["WGS 84",6378137,298.257223563,
LENGTHUNIT["metre",1]]],
PRIMEM["Greenwich",0,
ANGLEUNIT["degree",0.0174532925199433]],
ID["EPSG",4326]],
CONVERSION["UTM zone 19S",
METHOD["Transverse Mercator",
ID["EPSG",9807]],
PARAMETER["Latitude of natural origin",0,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8801]],
PARAMETER["Longitude of natural origin",-69,
ANGLEUNIT["degree",0.0174532925199433],
ID["EPSG",8802]],
PARAMETER["Scale factor at natural origin",0.9996,
SCALEUNIT["unity",1],
ID["EPSG",8805]],
PARAMETER["False easting",500000,
LENGTHUNIT["metre",1],
ID["EPSG",8806]],
PARAMETER["False northing",10000000,
LENGTHUNIT["metre",1],
ID["EPSG",8807]]],
CS[Cartesian,2],
AXIS["(E)",east,
ORDER[1],
LENGTHUNIT["metre",1]],
AXIS["(N)",north,
ORDER[2],
LENGTHUNIT["metre",1]],
USAGE[
SCOPE["Engineering survey, topographic mapping."],
AREA["Between 72°W and 66°W, southern hemisphere between 80°S and equator, onshore and offshore. Argentina. Bolivia. Brazil. Chile. Colombia. Peru."],
BBOX[-80,-72,0,-66]],
ID["EPSG",32719]]
Data axis to CRS axis mapping: 1,2
Origin = (619843.750000000000000,8895156.250000000000000)
Pixel Size = (2.229332773814519,-2.229332773814519)
Metadata:
AREA_OR_POINT=Area
TIFFTAG_COPYRIGHT=(C) Copyright 2021 DigitalGlobe, Inc., Westminster CO USA 80234
Image Structure Metadata:
COMPRESSION=DEFLATE
INTERLEAVE=PIXEL
LAYOUT=COG
Corner Coordinates:
Upper Left ( 619843.750, 8895156.250) ( 67d54'23.67"W, 9d59'35.02"S)
Lower Left ( 619843.750, 8889843.750) ( 67d54'23.10"W, 10d 2'27.97"S)
Upper Right ( 625156.250, 8895156.250) ( 67d51'29.21"W, 9d59'34.43"S)
Lower Right ( 625156.250, 8889843.750) ( 67d51'28.60"W, 10d 2'27.38"S)
Center ( 622500.000, 8892500.000) ( 67d52'56.14"W, 10d 1' 1.20"S)
Band 1 Block=512x512 Type=UInt16, ColorInterp=Gray
NoData Value=0
Overviews: 1192x1192, 596x596, 298x298
Band 2 Block=512x512 Type=UInt16, ColorInterp=Undefined
NoData Value=0
Overviews: 1192x1192, 596x596, 298x298
Band 3 Block=512x512 Type=UInt16, ColorInterp=Undefined
NoData Value=0
Overviews: 1192x1192, 596x596, 298x298
Band 4 Block=512x512 Type=UInt16, ColorInterp=Undefined
NoData Value=0
Overviews: 1192x1192, 596x596, 298x298
Band 5 Block=512x512 Type=UInt16, ColorInterp=Undefined
NoData Value=0
Overviews: 1192x1192, 596x596, 298x298
Band 6 Block=512x512 Type=UInt16, ColorInterp=Undefined
NoData Value=0
Overviews: 1192x1192, 596x596, 298x298
Band 7 Block=512x512 Type=UInt16, ColorInterp=Undefined
NoData Value=0
Overviews: 1192x1192, 596x596, 298x298
Band 8 Block=512x512 Type=UInt16, ColorInterp=Undefined
NoData Value=0
Overviews: 1192x1192, 596x596, 298x298
ogrinfo¶
This example lists information about a specified cloud mask GeoPackage (vector file).
ogrinfo /vsis3/maxar-ard-samples/rio_deforestation/19/300022033202/2013-08-28/1030010026C30800-cloud-mask.gpkg
Example using file from downloaded sample data:
ogrinfo /[root dir]/[subdir]sample-001-subset/10/120020223032/2019-07-12/1050010017603F00-cloud-mask.gpkg
INFO: Open of `/vsis3/maxar-ard-samples/rio_deforestation/19/300022033202/2013-08-28/1030010026C30800-cloud-mask.gpkg'
using driver `GPKG' successful.
1: 19_300022033202_1030010026C30800-cloud-mask (Multi Polygon)
Additional GDAL commands are described in the GDAL programs documentation.
Rasterio's "rio" command line tools¶
Rio is the Rasterio CLI. Use rio commands to get information about a file or to visualize a file.
Installation¶
Basic installation instructions for Rasterio can be found on the Rasterio Documentation Site.
Use rio to work with ARD Data¶
Once you install rasterio, you can use rio commands to access and work with ARD data. If the rio-viz subpackage is not installed, follow these instructions.
rio uses the same connection syntax as GDAL, so s3:// URLs should be changed to `/vsis3/.
rio info¶
rio info /vsis3/maxar-ard-samples/rio_deforestation/19/300022033202/2013-08-28/1030010026C30800-ms.tif
Example using downloaded sample data:
{"blockxsize": 512, "blockysize": 512, "bounds": [619843.75, 8889843.75, 625156.25, 8895156.25], "colorinterp": ["gray", "undefined", "undefined", "undefined", "undefined", "undefined", "undefined", "undefined"], "compress": "deflate", "count": 8, "crs": "EPSG:32719", "descriptions": [null, null, null, null, null, null, null, null], "driver": "GTiff", "dtype": "uint16", "height": 2383, "indexes": [1, 2, 3, 4, 5, 6, 7, 8], "interleave": "pixel", "lnglat": [-67.88226237156347, -10.017000640120592], "mask_flags": [["nodata"], ["nodata"], ["nodata"], ["nodata"], ["nodata"], ["nodata"], ["nodata"], ["nodata"]], "nodata": 0.0, "res": [2.2293327738145194, 2.2293327738145194], "shape": [2383, 2383], "tiled": true, "transform": [2.2293327738145194, 0.0, 619843.75, 0.0, -2.2293327738145194, 8895156.25, 0.0, 0.0, 1.0], "units": [null, null, null, null, null, null, null, null], "width": 2383}
rio viz¶
The rio viz command line tool provides a visual browser to explore ARD image tiles.
rio viz /vsis3/maxar-ard-samples/sample-002/13/031133231113/2021-01-16/10300100B3841C00-visual.tif

Example using the downloaded sample data:
rio viz /[root dir]/[subdir]/sample-001-subset/10/120020223032/2019-07-12/1050010017603F00-visual.tif
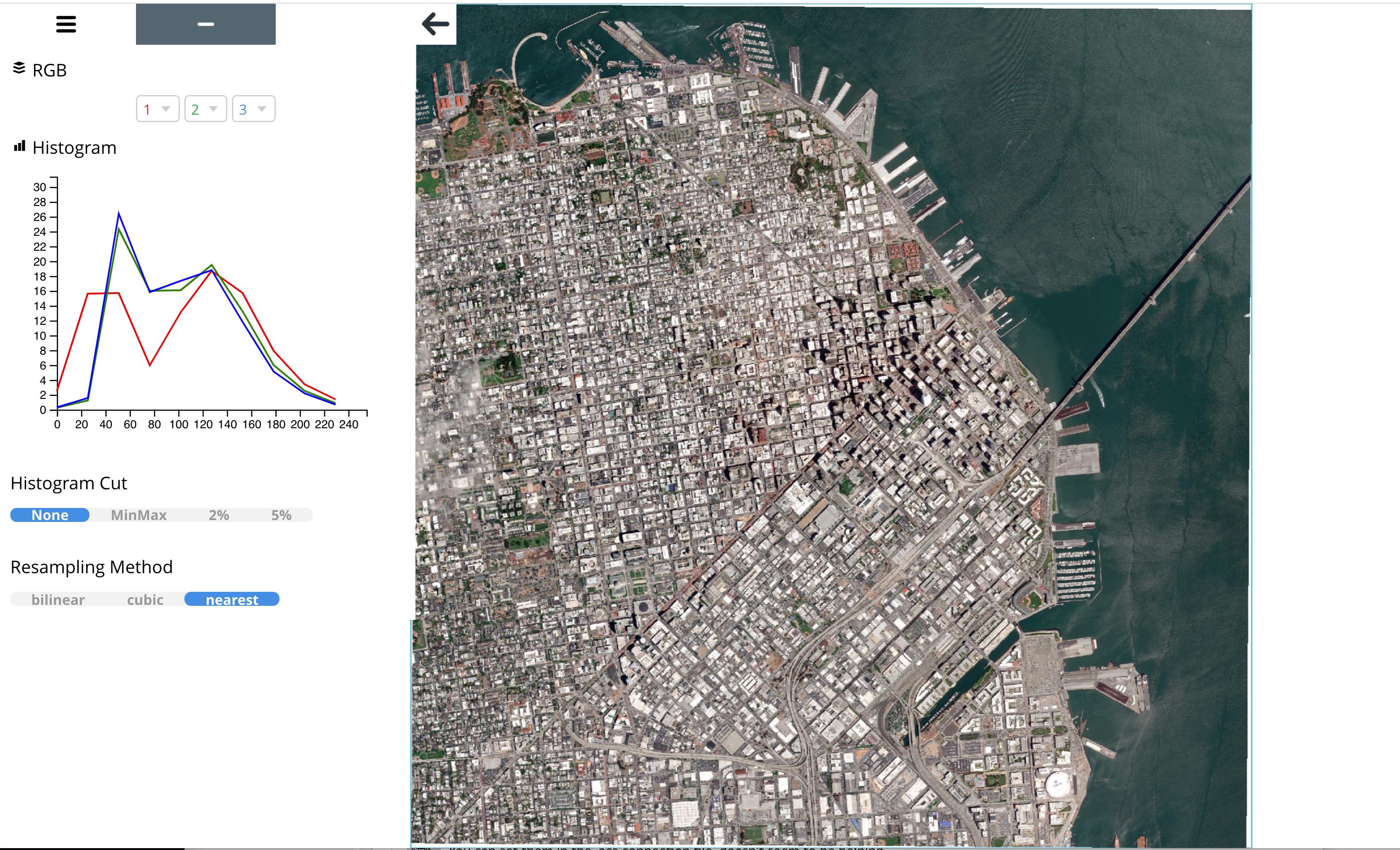
Rio viz can also read the ARD STAC Items.
Example:
rio viz https://maxar-ard-samples.s3.amazonaws.com/sample-002/13/031133231113/2021-01-16/10300100B3841C00.json --reader rio_tiler.io.STACReader